Introduction
The purpose of StructureFinder is to find crystallograpgic files such as Crystallographic Information Files (.cif) and SHELX (.res) files. StructureFinder indexes all .cif and/or .res files below a certain directory(s) and makes them searchable. The intention is not to bring the files in order or have a static database. It only reflects the order of files in the file system.
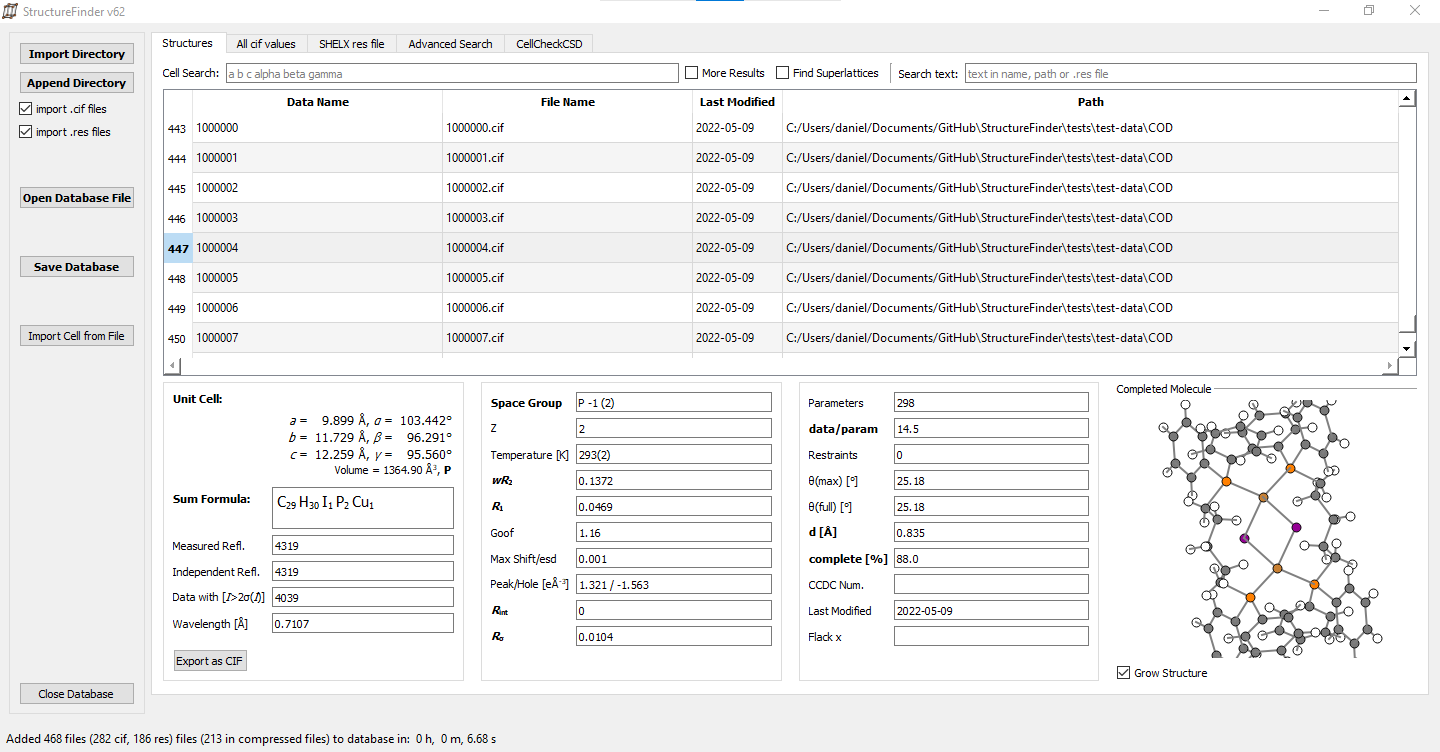
The StructureFinder main window.
In the main tab, you can import CIF and SHELX .res files to a database and you can save this database to a file for later usage.
Selecting a certain row of the database shows the unit cell, the residuals and the asymmetric unit.
Basic search options for unit cells and text is also available. The cell search takes six parameters a, b, c, α, β, γ. The search is unsharp so 10 10 10 90 90 90 would find the same cell as 10.00 10.00 10.00 90.00 90.00 90.00.
The algorithm is a combination of a cell comparison by volume first (for speed) and subsequent matching. The lattice matching implementation was made by the pymatgen project. The tolerances for the cell search are:
regular volume: ±3 %, length: 0.06 Å, angle: 1.0°
more results option volume: ±9 %, length: 0.2 Å, angle: 2.0°
The text search field searches in the directory, name and .res file text data. You can concatenate words with ? and *. For example foo*bar means ‘foo[any text]bar’.
Pro tip: Double click on the unit cell to copy the cell to the clip board.
Customizing columns
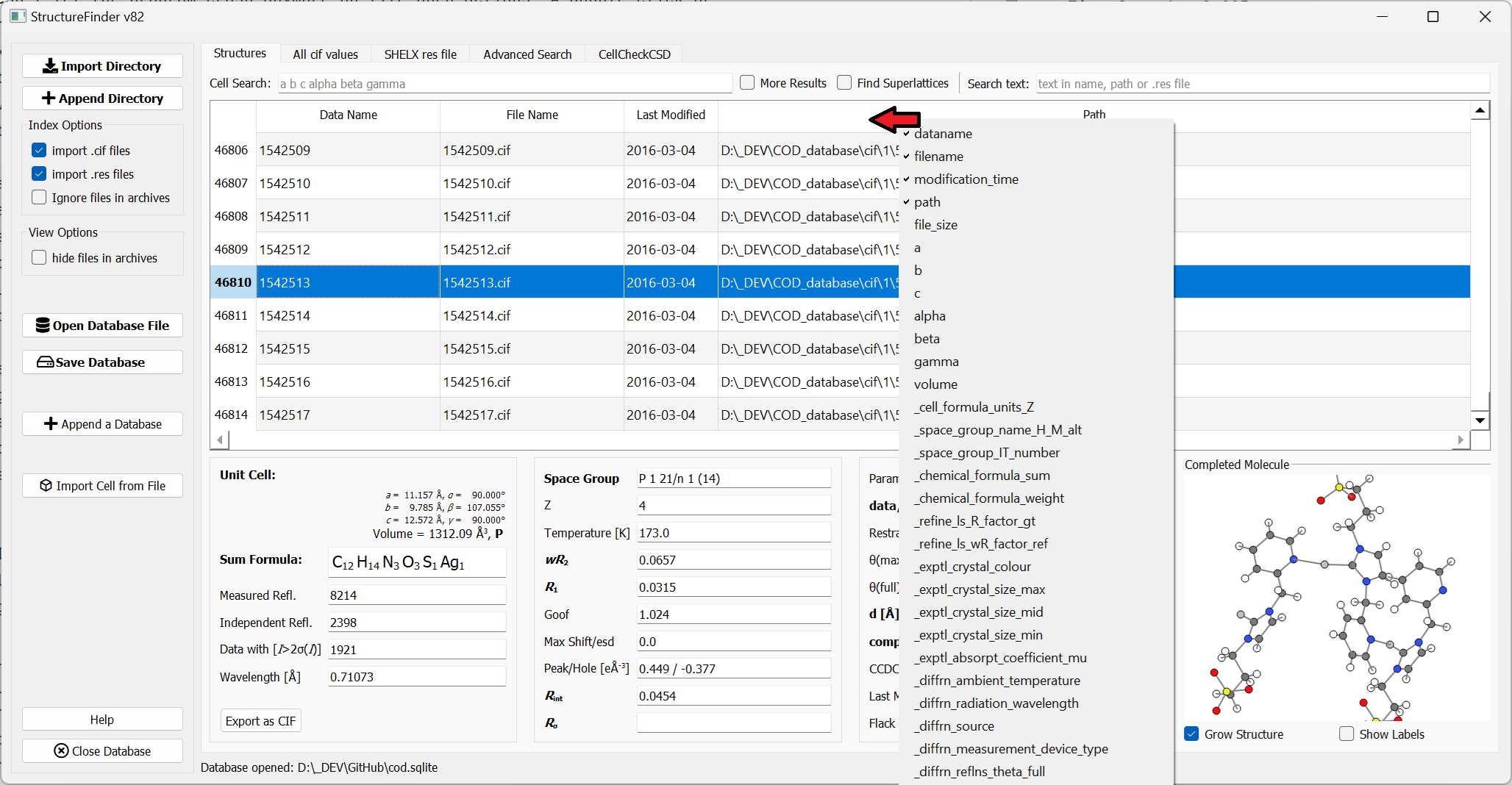
List of available columns.
The columns shown in the table can be changed by right-click on the table header. The selcted columns will be saved during close of the application.
All CIF values tab
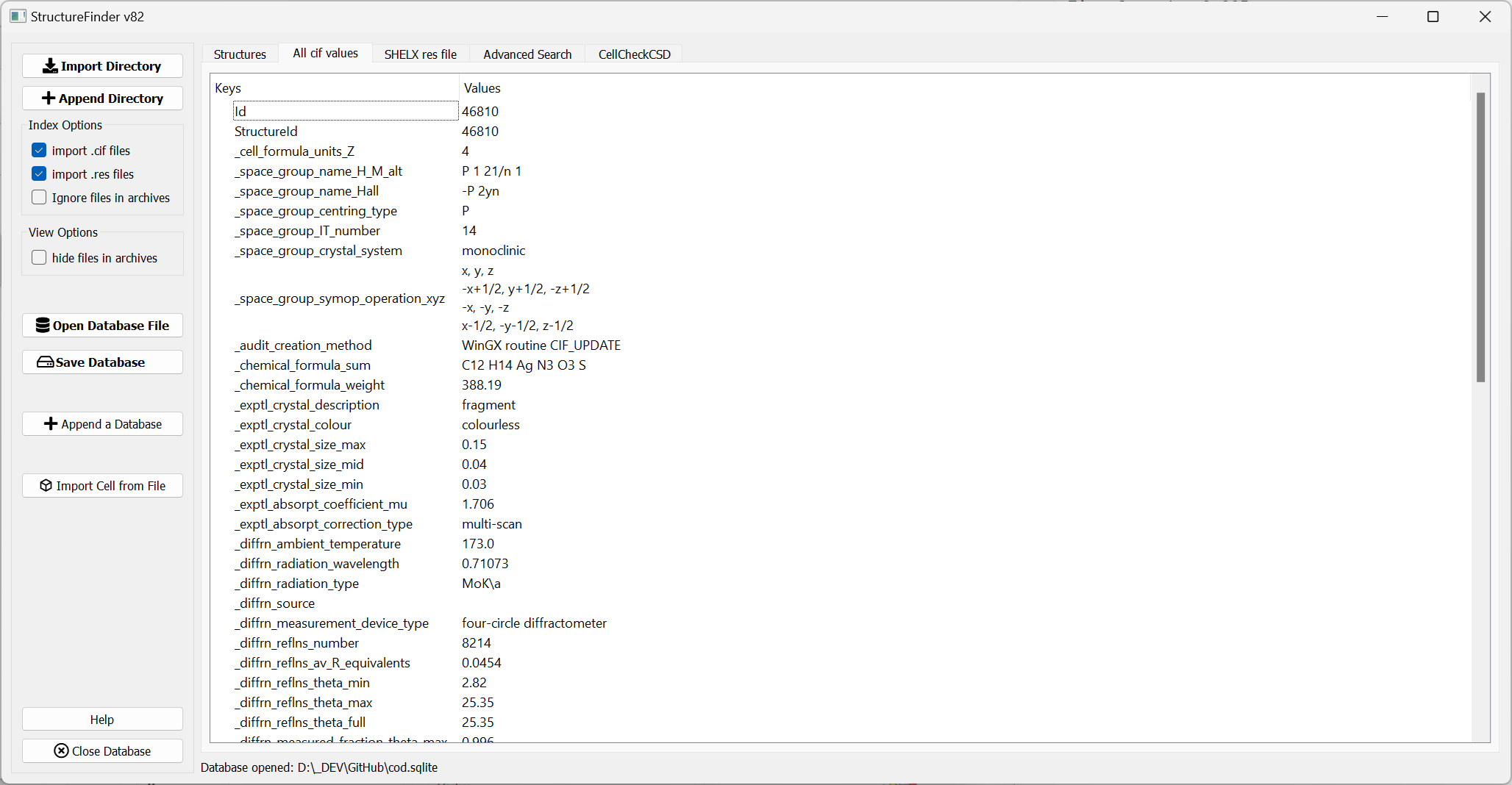
List of all CIF values for one structure.
The ‘All CIF values’ tab shows all cif values available in the database. These are not necessarily all but most values from the cif file.
Advanced Search
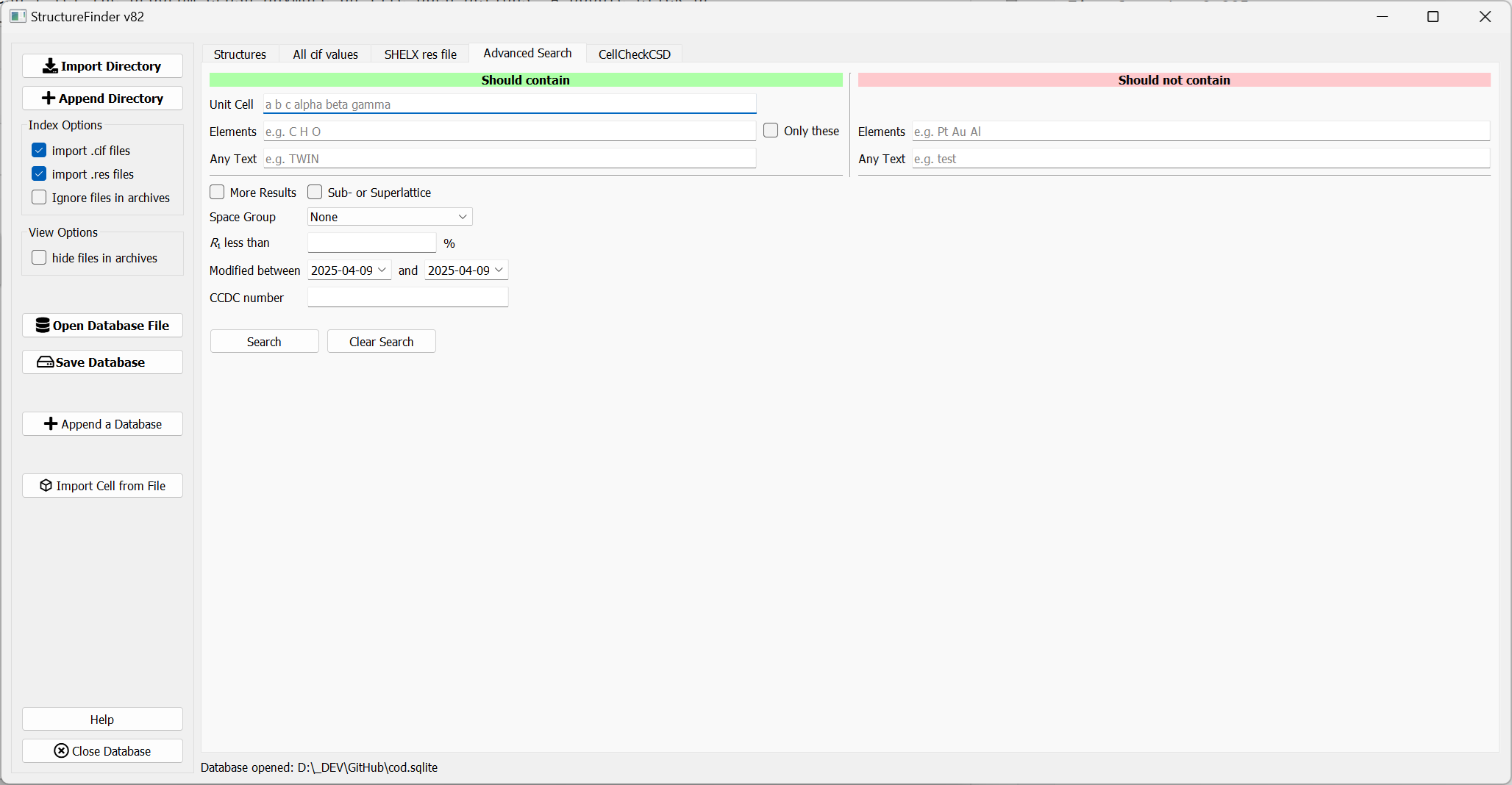
Advanced search tab.
The ‘Advanced Search’ tab allows you to search for several options at a time and also allows to exclude parameters. I will add more options in the future and I am open for suggestions for more search options.
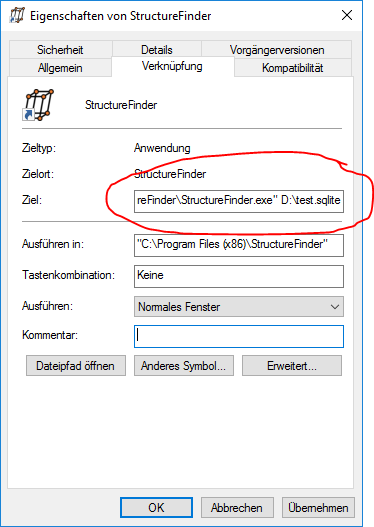
Open Database Automatically
If you want to open the same database file with the Windows version, you can add the database file as command line parameter in the start menu shortcut:
Indexing files
The ‘Import Directory’ button starts indexing of .cif or .res files below the selected directory recursively, depending on which file ending is enabled. It will scan all subdirectories for .cif/.res files as well as .zip files containing .cif files. The time for indexing mostly depends on the speed of your hard drive. Scanning a complete 256 GB SSD takes about 50s. An ten years old complete file server with over 100 user directories and 20.000 crystal structures can take an hour.
Be aware that the indexing on a network drive is very slow!
Installation
Windows
The windows version installs like any other Windows installer.
Linux / Mac / Windows
Since version 73, there is a pypi package for installation in a Python environment. Do the following steps in order to install and run StructureFinder in any Python environment:
>> python -m venv venv <-- creates a virtual environment
>> source venv/bin/activate (Windows: venv\Scripts\activate.bat) <-- Activates the environment
>> pip install structurefinder
>> strf or strf_cmd or strf_web
For the next start, only
>> source venv/bin/activate (Windows: venv\Scripts\activate.bat)
>> strf or strf_cmd or strf_web
is necessary.
Command Line file Indexer
With the strf_cmd command, you can index directories without a graphical user interface.
The options -d and -e can be given multiple times like -d /foo -d /bar.
$ strf_cmd
usage: strf_cmd.py [-h] [-d "directory"] [-e "directory"]
[-o "sqlite file name"] [-c] [-r] [--delete]
[-f "unit cell"] [-m "sqlite file name"] [-na]
Command line version 82 of StructureFinder to collect .cif/.res files to a
database. StructureFinder will search for cif files in the given directory(s)
recursively. (Either -c, -r or both options must be active!)
options:
-h, --help show this help message and exit
-d "directory" Directory(s) where cif files are located.
-e "directory" Directory names to be excluded from the file search.
Default is: ['BrukerShelXlesaves',
'autostructure_private', '.OLEX', 'TEMP', 'ROOT',
'olex', '__private__', 'dsrsaves', 'shelXlesaves',
'Papierkorb', 'Recycle.Bin', 'TMP'] Modifying -e
option discards the default.
-o "sqlite file name"
Name of the output database file. Default:
"./structuredb.sqlite" Also used for the commandline
search (-f option).
-c Add .cif files (crystallographic information file) to
the database.
-r Add SHELX .res files to the database.
--delete Delete and do not append to previous database.
-f "unit cell" Search for the specified unit cell. The cell values
have to be enclosed in brackets.
-m "sqlite file name"
Merges a database file into the file of '-o' option.
Only -o is allowed in addition.
-na Disables the collection of files in .zip, .tar.gz,
.tar.bz2, .tgz, .7z archives.
Indexing Example
Creates the file structuredb.sqlite in the current directory:
Collecting .res and .cif files below D:\Github\Structurefinder.
Added 253 files, (249 .cif, 4 .res files), (209 in compressed archives) to database in: 0 h, 0 m, 0.87 s
---------------------
Total 253 cif/res files in 'D:\GitHub\StructureFinder\structuredb.sqlite'.
The command line version always appends all data to an already existing database in the current working directory. It will not append the date with the –delete option.
Database Format
The database format is just plain sqlite (http://www.sqlite.org/). You can view the database structure with the sqlitebrowser (http://sqlitebrowser.org/) for example.
CSD search
StructureFinder is able to search for unit cells in the CSD with the CellCheckCSD program. As soon as CellCheckCSD is installed, you can search the CSD. Double-Click on a result row to get the detailed structure page.
Web Interface
Instead of the regular user interface, you can run StructureFinder as web service. First, create a database with ./strf_cmd This can be automated with a cron job to do it regularly.
usage: strf_web [-h] [-n HOST] [-p PORT] [-f DBFILENAME] [-d]
StructureFinder Web Server v73
options:
-h, --help show this help message and exit
-n HOST, --host HOST
-p PORT, --port PORT
-f DBFILENAME, --dbfile DBFILENAME
-d, --download Shows a download link in the page bottom
The easiest way is to run ‘strf_web’ from a directory with a database file: strf_web -f structuredb.sqlite
Be aware that running a web server has security implications. Do not expose this server to the internet unless you know what you are doing!
The web site should look like this after clicking on a table row:
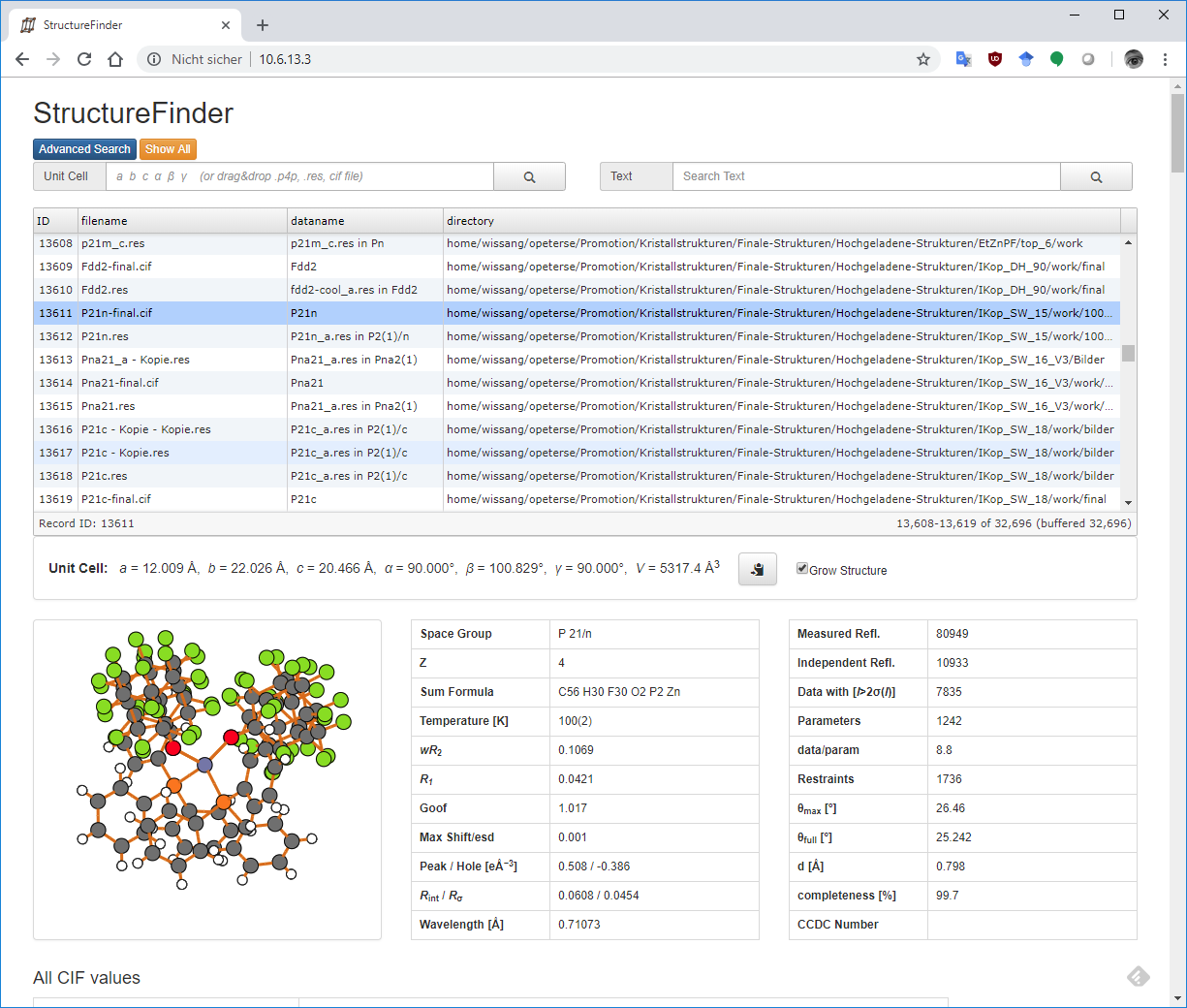
The StructureFinder web interface.