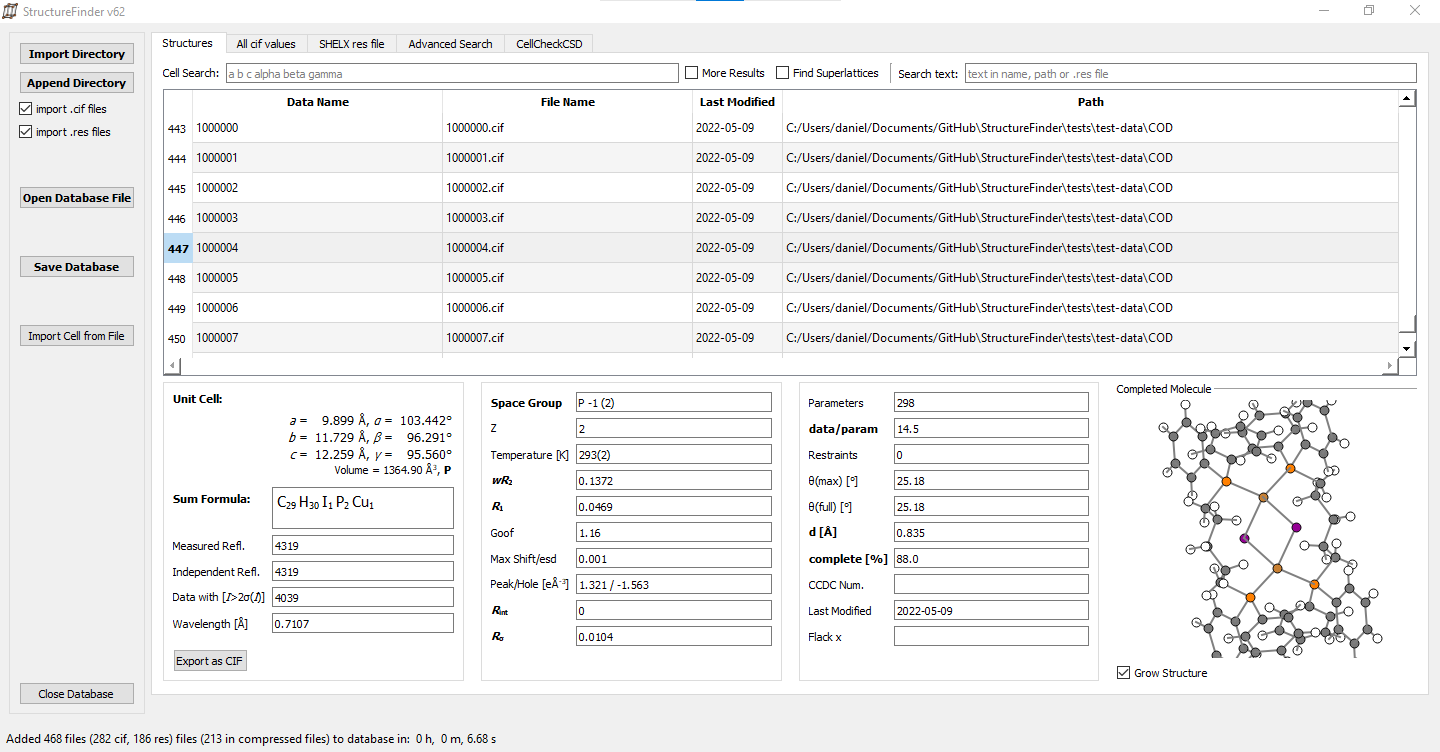
StructureFinder
The purpose of StructureFinder is to find crystallograpgic files such as Crystallographic Information Files (.cif) and SHELX (.res) files. StructureFinder indexes all .cif and/or .res files below a certain directory(s) and makes them searchable. The intention is not to bring the files in order or have a static database. It only reflects the order on the file system.
Downloads
| Current release | ||
|---|---|---|
| StructureFinder-setup-x64-v87.exe | 2025-11-24 | Windows 10 and up |
There is also a pypi package. See documentation for details.
Documentation
Recent Changes
See last changes in StructureFinder
Screenshots
Source Code
You can find the source code of StructureFinder at Github. Contributions are welcome!
License
StructureFinder is free software and licensed under the beerware license.