Disordered Structure Refinement (DSR)
The refinement of highly disordered solvents and moieties with SHELXL is an often slow or even difficult task. Therefore, these voids frequently are "squeezed out" of the X-ray data. This practice is often not necessary. To help with the routine work of refining disordered solvents the program DSR was developed in Python.
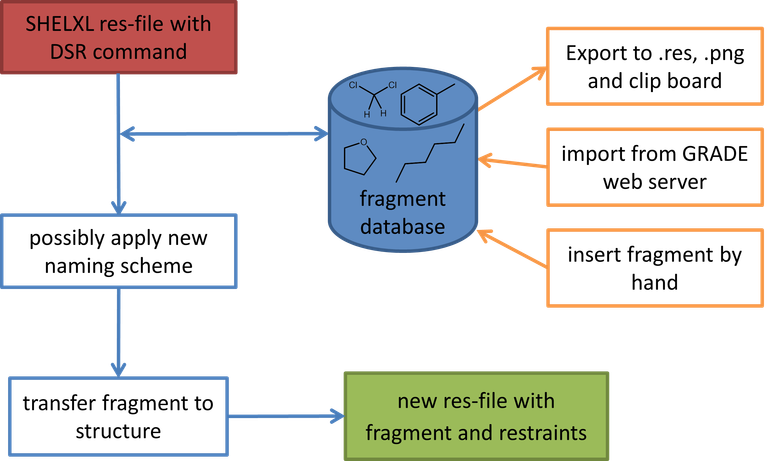
The program-package consists of a simple text-database with fragments of molecules and the DSR program itself. It acts as a preprocessor for SHELXL res-files. The user inserts a special command in the SHELXL .res file and the DSR program reads this information to place a molecular fragment with the desired atoms on the position of the target atoms or Q-peaks, respectively. Bond restraints are applied from the database to the fragment. DFIX restraints can be generated automatically, if desired. Ready to use fragments with sensible restraints can be imported from http://grade.globalphasing.org.
Please report bugs or annoying problems to dkratzert@gmx.de. I can only fix bugs that are known to me. Also wishes for new fragments or program features are very welcome!
You may cite DSR (or FragmentDB) as
D. Kratzert, I. Krossing, J. Appl. Cryst. 2018, 51, 928-934. doi: 10.1107/S1600576718004508 (direct download)
D. Kratzert, J.J. Holstein, I. Krossing, J. Appl. Cryst. 2015, 48, 933-938. doi:10.1107/S1600576715005580 (download)
DSR GUI
You find a graphical user interface for DSR in ShelXle. Just select three source and three target atoms each, and you can transfer the desired fragment from DSR into your structure in ShelXle. The edit window makes it easy to modify/create fragments. See a YouTube video how it works. You can also use DSR on the command line.
Known bugs
-
SHELXL-2019/1 has a bug where the combination of RESI and SAME leads to errors like
** SAME/SADI sfac type mismatch for C1_1 C2_1 F2_1 F9_1 **
** SAME/SADI sfac type mismatch for...
[and several more lines]The problem is solved in SHELXL-2019/2, which you can get on the SHELX website.
-
For Linux and Mac users: The update from DSR for python 2.7 (version 236 and below) to DSR for Python 3 needs some manual work. You should uninstall the old version and make sure the files dsr.sh, dsr.pth, dsr-mac and dsr-linux are removed from /etc/profile.d/ and/or /etc/paths.d/.
The command 'dsr -u' for updates doesn't work anymore!
Documentation
Downloads
| Current release | ||
|---|---|---|
| DSR-setup-242.exe | 2024-10-07 | Windows 10 and up |
| manual installation | Other non-Windows systems | |
Alternatively, you can install DSR from PyPI:
Install Python version >= 3.9, and run 'pip install dsr-shelx'.
(The project name dsr was already taken).
The PyPI package has no external dependencies, so it can run on any system that supports Python3.
Recent Changes
changelogMiscelanious
- You prefer Olex2? Then you might want to use the FragmentDB plugin. It is a port of DSR to Olex2.
- You just need a parser for SHELX files? Then you might be interested in my ShelXFile project.
Some molecular fragments are taken from the "Idealized Molecular Geometry
Library" of Ilia A. Guzei
(I. A. Guzei, J. Appl. Cryst. 2014, 47, 806-809).
DSR uses the following Python libraries:
Fredrik Johansson et.al. mpmath: a Python library for arbitrary-precision floating-point arithmetic (version 0.18), December 2013, http://mpmath.org/.
A. Hagberg, D. Schult, P. Swart, NetworkX: package for the creation, manipulation, and study of complex networks (version 1.8.1) 2013, http://networkx.github.io/.
Source Code
You can find the source code of DSR at Github.
License
DSR is free software and licensed under the beerware license.